ordmmd
Compute multiple minimum degree ordering
Syntax
[perm,invp,nofsub]=ordmmd(xadj,iadj,n)
Arguments
- n
a 1-by-1 matrix of doubles, integer value, the number of equations
- xadj
(n+1)-by-1 matrix of doubles, integer value, pointer to the rows of A
- iadj
nnz-by-1 matrix of doubles, integer value, the row indices of A
- perm
n-by-1 matrix of doubles, integer value, the minimum degree ordering
- invp
n-by-1 matrix of doubles, integer value, the inverse of perm
- nofsub
1-by-1 matrix of doubles, integer value, an upper bound on the number of nonzero subscripts for the compressed storage scheme
Description
The minimum degree algorithm is used to permute the rows and columns of a symmetric sparse matrix before applying the Cholesky decomposition. This algorithm reduces the number of non-zeros in the Cholesky factor.
Given a n-by-n real symmetric sparse square matrix A,
the Cholesky factor U will typically suffer "fill in", that is have more
non-zeros than the upper triangle of A.
We seek a permutation matrix P, so that the matrix P'*A*P,
which is also symmetric, has the least possible fill in its Cholesky factor.
Examples
In the following example, we compute an ordering for a symmetric
sparse matrix.
We use the sp2adj function to compute the adjacency structure.
A = [ 4. 1. 2. 0.5 2. 1. 0.5 0. 0. 0. 2. 0. 3. 0. 0. 0.5 0. 0. 5./8. 0. 2. 0. 0. 0. 16. ]; A = sparse(A); [xadj,iadj,val]=sp2adj(A); n = size(A,"r"); [perm,invp,nofsub]=ordmmd(xadj,iadj,n)
In the following example, we compute an ordering for a symmetric
sparse matrix.
We check that invp is the inverse of perm.
A = [ 0., 0., 0., 2., 0., 0., 2., 0., 2., 0., 0. ; 0., 0., 4., 0., 0., 0., 0., 0., 0., 0., 0. ; 0., 4., 0., 0., 0., 0., 0., 0., 0., 0., 0. ; 2., 0., 0., 0., 0., 0., 2., 0., 2., 0., 0. ; 0., 0., 0., 0. , 0., 0., 0., 0., 0., 0., 4. ; 0., 0., 0., 0., 0., 0., 0., 3., 0., 3., 0. ; 2., 0., 0., 2., 0., 0., 0., 0., 2., 0., 0. ; 0., 0., 0., 0., 0., 3., 0., 0., 0., 3., 0. ; 2., 0., 0., 2., 0., 0., 2., 0., 0., 0., 0. ; 0., 0., 0., 0., 0., 3., 0., 3., 0., 0., 0. ; 0., 0., 0., 0., 4., 0., 0., 0., 0., 0., 0.]; n=size(A,1); A=sparse(A); [xadj,adjncy,anz]=sp2adj(A); [perm,invp,nofsub]=ordmmd(xadj,adjncy,n); perm(invp)
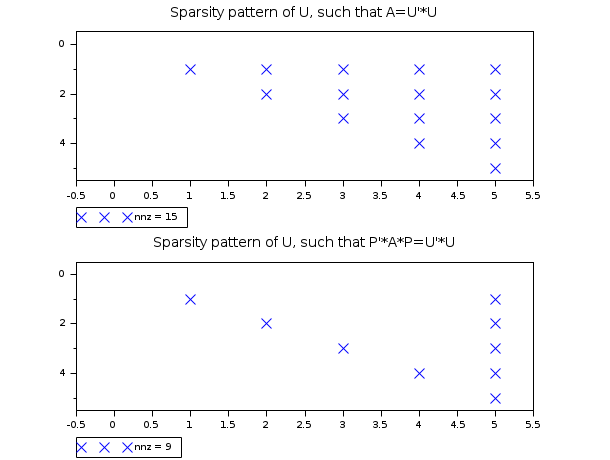
In the following example, we compare the sparsity pattern of the Cholesky
factor of a matrix A and the matrix P'*A*P.
See p. 3, "Chapter 1: Introduction" in
"Computer Solution of Large Sparse Positive Definite Systems".
We see that the number of nonzeros in the Cholesky decomposition is
15, while the matrix P'*A*P has a Cholesky decomposition with
9 nonzeros.
A = [ 4. 1. 2. 0.5 2. 1. 0.5 0. 0. 0. 2. 0. 3. 0. 0. 0.5 0. 0. 5./8. 0. 2. 0. 0. 0. 16. ]; A = sparse(A); // See the sparsity pattern of the Cholesky factors of A U = sparse(chol(full(A))); scf(); subplot(2,1,1); PlotSparse(U,"x"); xtitle("Sparsity pattern of U, such that A=U''*U"); // Get the adjacency structure [xadj,iadj,val]=sp2adj(A); // Compute multiple minimum degree ordering n = size(A,"r"); [perm,invp,nofsub]=ordmmd(xadj,iadj,n); // Convert the permutation vector into matrix. P=spzeros(n,n); for i=1:n P(perm(i),i)=1; end // See the sparsity pattern of the Cholesky factors // of P'*A*P U = sparse(chol(full(P'*A*P))); subplot(2,1,2); PlotSparse(U,"x"); xtitle("Sparsity pattern of U, such that P''*A*P=U''*U");
Implementation notes
This function is based on "ordmmd.f" a source code (1994) by Esmond G. Ng and Barry W. Peyton from Mathematical Sciences Section, Oak Ridge National Laboratory. The algorithm is based on the genmmd routine by Joseph W.H. Liu from the SPARSPAK library.
Bibliography
"Minimum degree algorithm", Wikipedia contributors, Wikipedia, The Free Encyclopedia. http://en.wikipedia.org/wiki/Minimum_degree_algorithm
"A new release of SPARSPAK: the Waterloo sparse matrix package", Alan George and Esmond Ng. 1984. SIGNUM Newsl. 19, 4 (October 1984), 9-13.
"Computer Solution of Large Sparse Positive Definite Systems" by Alan George and Joseph Liu, Prentice-Hall, Inc. Englewood Cliffs, New Jersey, 1981
"Implementation of Lipsol in Scilab", Rubio Scola, 1997, INRIA, No 0215
See also
| Report an issue | ||
| << chsolve | Matrizes Esparsas | Interfaces com UMFPACK (sparse) >> |