Please note that the recommended version of Scilab is 2026.0.1. This page might be outdated.
See the recommended documentation of this function
h5write
Create a dataset (if it does not exist) and write the data
Syntax
h5write(obj, name, data [, targetType], [, start, count [, stride [, block]]]) h5write(filename, name, data [, targetType], [, start, count [, stride [, block]]])
Arguments
- obj
a H5Object
- name
a string giving the path to the dataset
- data
a Scilab data
- targetType
a string giving the type of the target
- start
a row of doubles containing the start of the hyperslab
- count
a row of doubles containing the count of the hyperslab
- stride
a row of doubles containing the stride of the hyperslab
- block
a row of doubles containing the block of the hyperslab
- filename
a string giving the filename
Description
Create a new named dataset (if it does not exist) based on the Scilab data passed as argument.
The target HDF5 type can be chosen in the list available in the HDF5 manual. Examples of this HDF5 type are "H5T_MIPS_U32" or "H5T_STD_B64BE", but shortcuts as "MIPS_U32" or "STD_B64BE" can also be used.
It is possible to make an hyperslab selection on the data.
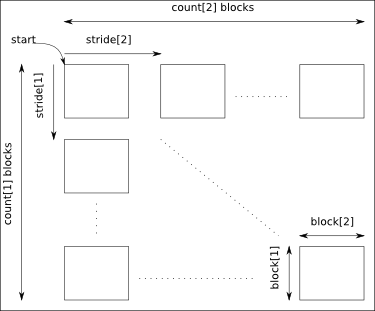
The arguments start, count, stride and block must have a size equal to the number of dimensions of the data:
- start: gives the coordinates in the data where to start the selection.
- count: gives the number of selected blocks in each dimension.
- stride: gives the shift between two consecutives blocks in each dimension. Take care that the stride must be greater than the corresponding block dimension.
- block: gives the block dimensions.
If the target type is "H5T_STD_REF_OBJ", the data matrix must be a string matrix containing the absolute paths of group or datasets.
Examples
x = matrix(1:20, 4, 5); save(TMPDIR + "/x.sod", "x"); // SOD files are HDF5 ones // Open the created file a = h5open(TMPDIR + "/x.sod"); // We want to add a new dataset named "y" y = uint32(matrix(10:30, 7, 3)); h5write(a, "y", y); // We add a dataset "z" in using MIPS representation h5write(a, "z", y, "H5T_MIPS_U32"); // Now we make an hyperslab selection x = uint32(matrix(1:(11*17), 11, 17)); h5write(a, "t", x, [1 2], [2 4], [5 3], [3 2]); //Now check that all is ok x, a("/t").data' // We have finished so we free all the resources h5close(a);
History
| Version | Description |
| 5.5.0 | HDF5 module introduced. |
| Report an issue | ||
| << h5umount | HDF5 Management | h5writeattr >> |